Antisense oligonucleotides are typically 15-20 bases in length. There are many factors to consider when designing ASOs. Sequences too long are not easily absorbed by cells, while too short would lack specificity to the target RNA.
Target Structure
Inside cells, RNA folds into secondary structures and form complexes with ribonucleoproteins into tertiary structures. These higher order structures can prevent the ASO from hybridizing to their theoretical target sequence. So when selecting a suitable target sequence, the predicted secondary and tertiary structures should be considered, and in vivo binding studies should be performed to confirm target-binding.
Nuclease Stability
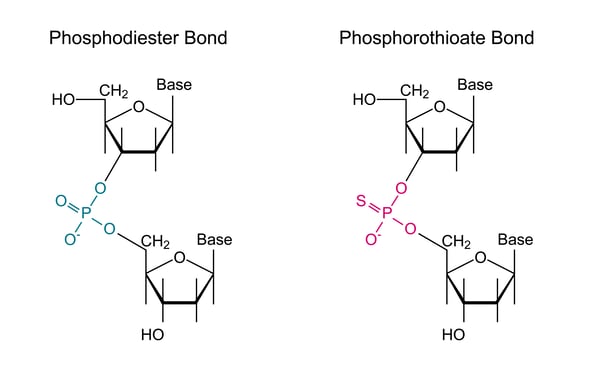
Both endonucleases and exonucleases can cause degradation in vivo. All ASOs are chemically modified to resist nuclease degradation. Phosphorothiothioate modifications along the backbone of the ASO are introduced to resist nuclease degradation.
Secondary Structure and Dimers
When designing the sequence, base self-pairing in the antisense oligonucleotide and formation of ASO dimers should be avoided. Both self-pairing and dimer formation can hinder the oligonucleotide’s ability to effectively bind to the target sequence.
The GC content of ASOs should be around 60%-65%. High GC content can cause self-aggregation and reduce the availability of ASOs to the target sequence, while low GC content may decrease affinity to the target.
Functional Motifs
The motifs CCAC, TCCC, ACTC, GCCA, and CTCT were associated with enhanced efficiency, while GGGG, ACTG, AAA, TAA and these motifs reduced antisense activity.
Off-Target
The final unmodified antisense oligonucleotide sequence should be searched for BLAST to check for any off-target hybridization.